Detect outliers from hdbscan for large data
outlier_hdbscan.RdObtain aggreagted GLOSH outlier scores based on hdbscan
outlier_hdbscan(mat, k, sampleSize, nEpochs, distMethod = "euclidean", seed = 1, nproc = 1, distFunc)
Arguments
| mat | (numeric matrix) data matrix |
|---|---|
| k | (pos int) Minimum size of clusters for hdbscan |
| sampleSize | (pos int) Size of the sample |
| nEpochs | (pos int) Number of samples |
| distMethod | (string) Method of compute distance matrix. Default is 'euclidean' |
| seed | (pos int) seed |
| nproc | (pos int) Number of parallel processses to use via forking |
| distFunc | 'fun' argument for 'parallelDist::parDist' when distMethod is "custom" |
Value
A vector of outlier scores
Examples
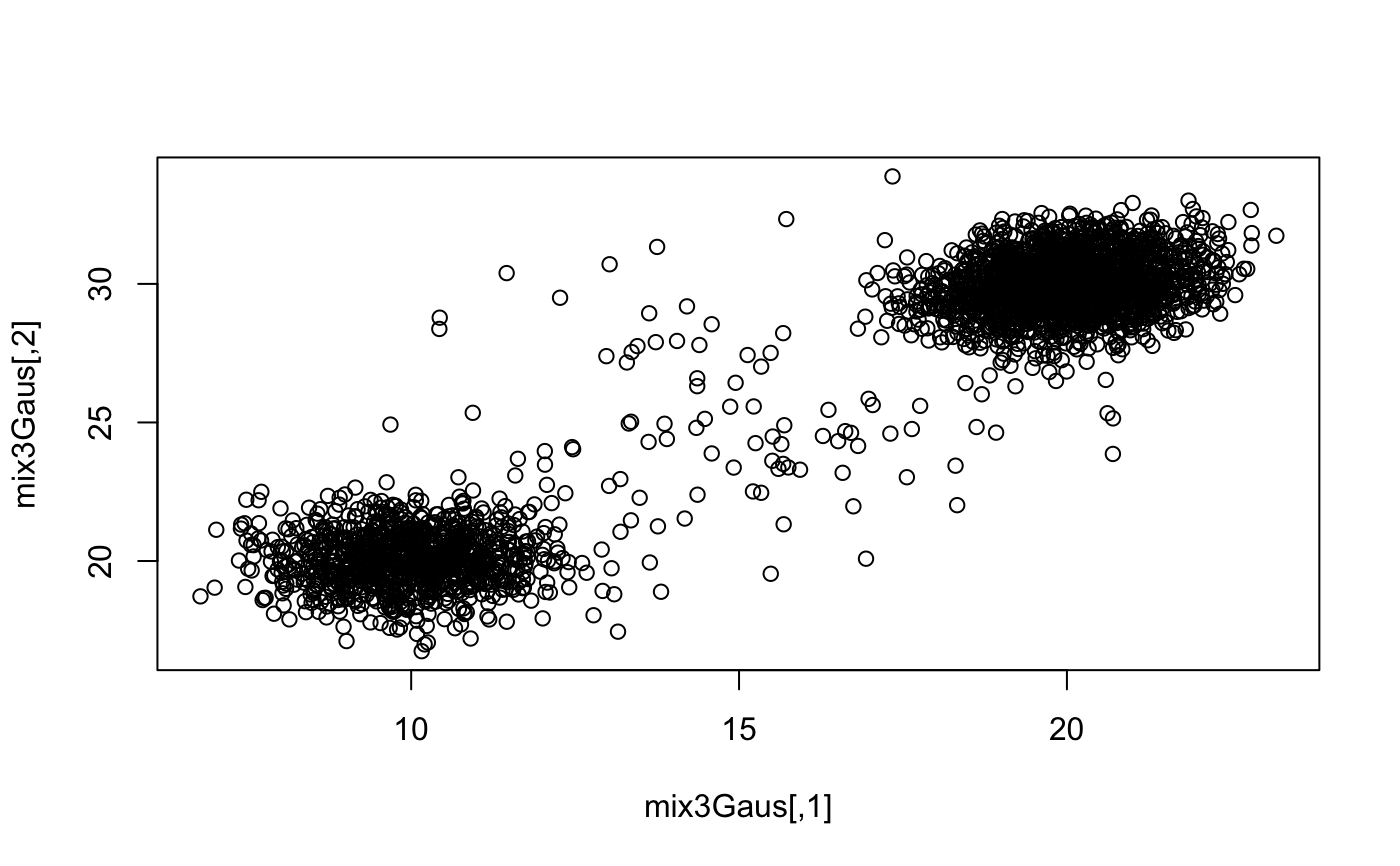
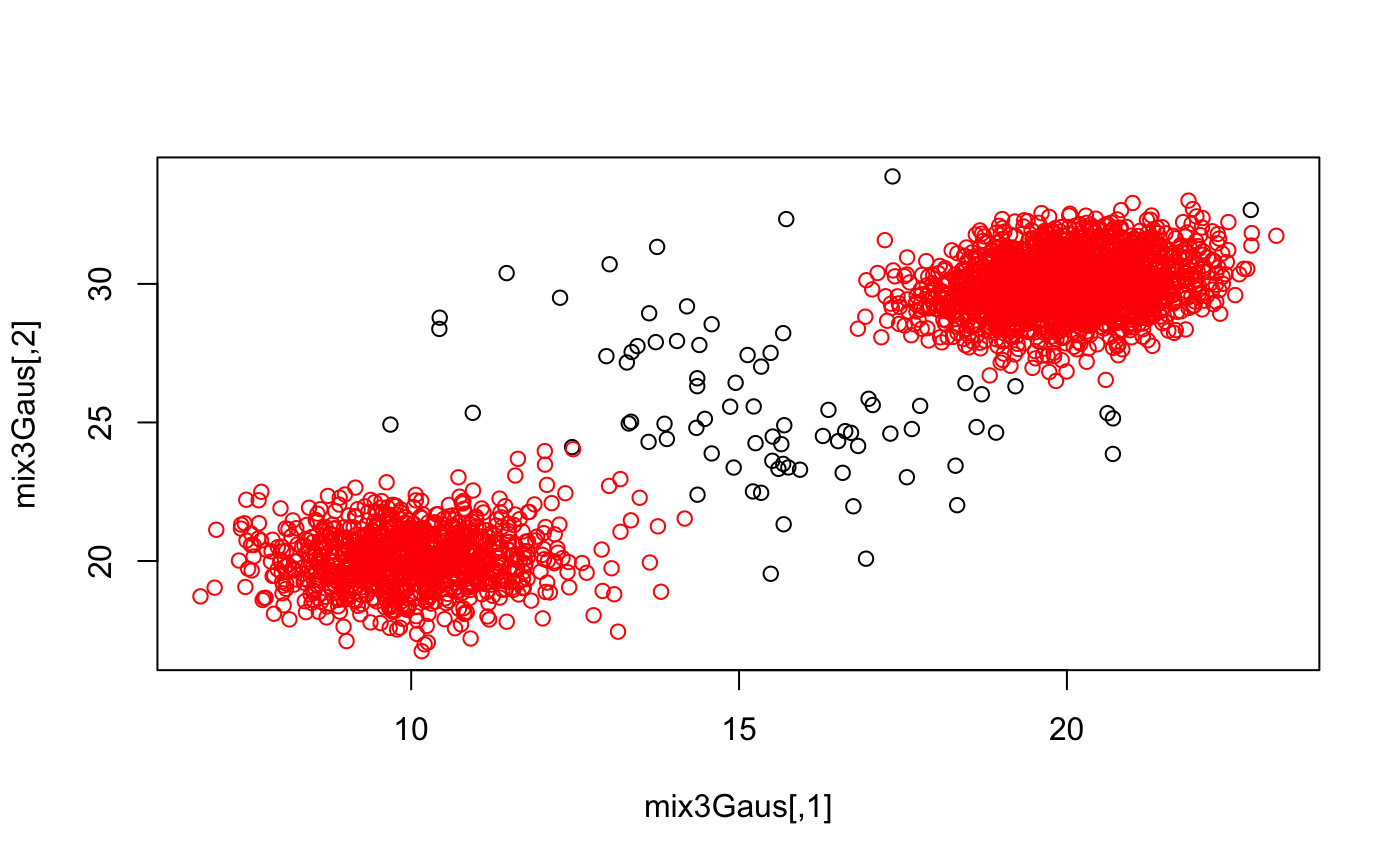
set.seed(1) mix3Gaus <- rbind( mvtnorm::rmvnorm(1e3, mean = c(10, 20)) , mvtnorm::rmvnorm( 2e3 , mean = c(20, 30) , sigma = matrix(c(1, 0.2, 0.2, 1), ncol = 2)) , mvtnorm::rmvnorm(100, mean = c(15, 25), sigma = diag(6, 2)) ) mix3Gaus <- mix3Gaus[sample(nrow(mix3Gaus)), ] outScore <- outlier_hdbscan(mat = mix3Gaus , k = 100 , sampleSize = 1e3 , nEpochs = 1e2 ) plot(density(outScore))plot(mix3Gaus)