Sample points based on distance matrix
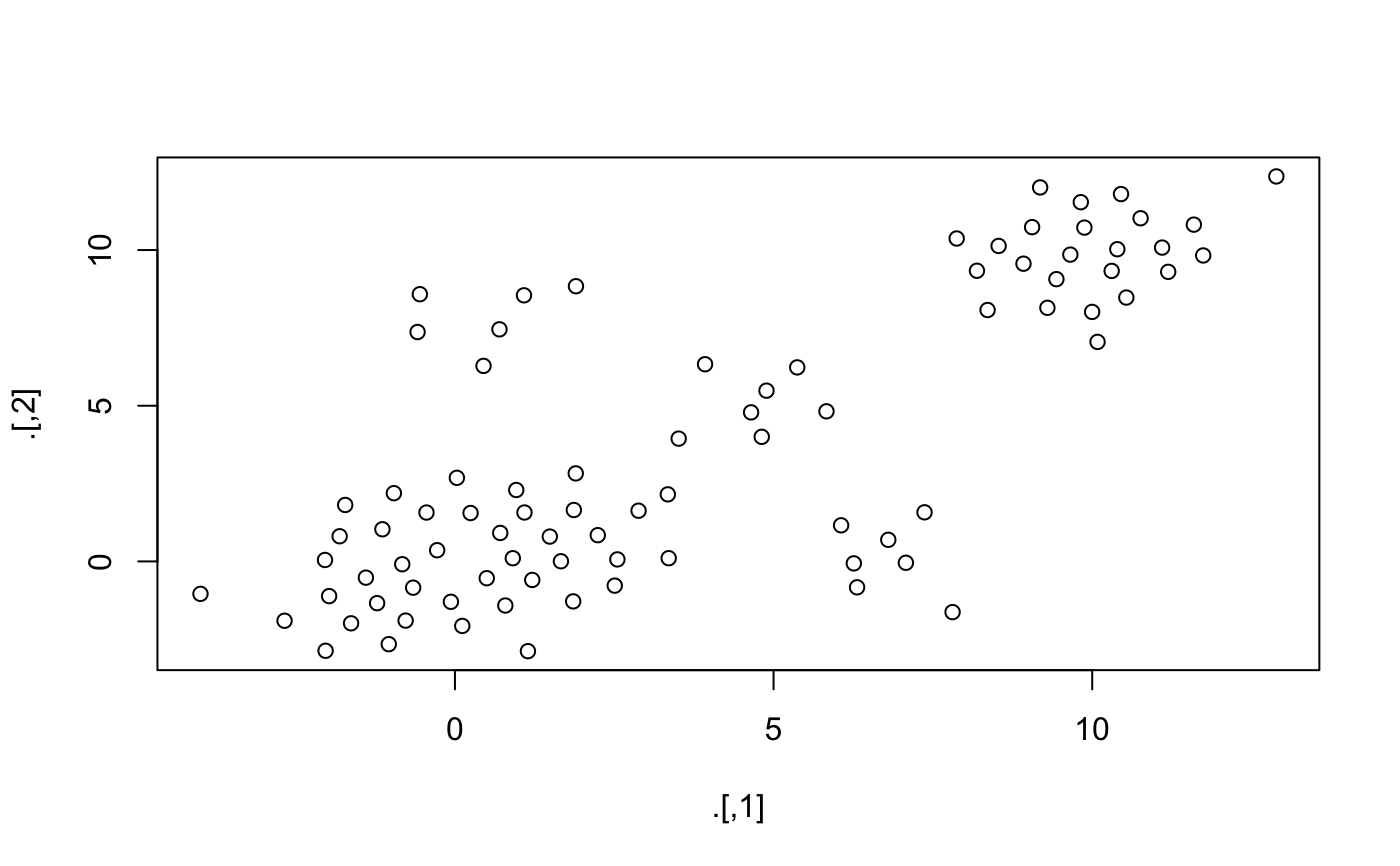
condense.RdSample points based on distance matrix so that distance between any pair is not less than a fixed radius. This will not preserve density, but does preserve local outliers.
condense(d, radius, seed = 1, ...)
Arguments
| d | object of class 'dist' |
|---|---|
| radius | (numeric) radius around a point |
| seed | (positive integer) A seed. Default is 1. |
| ... | Arguments for |
Value
Index of condensed points
Examples
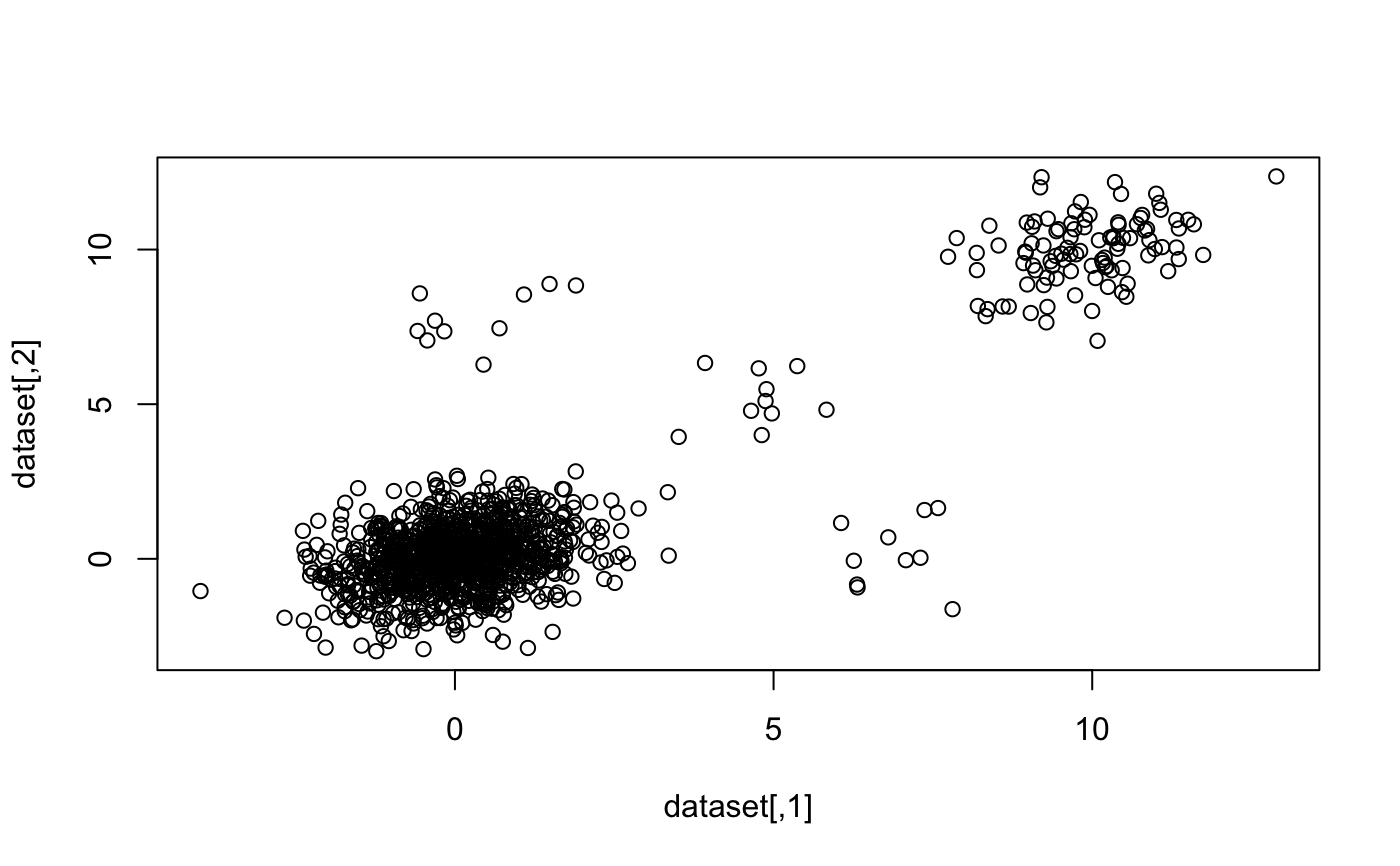
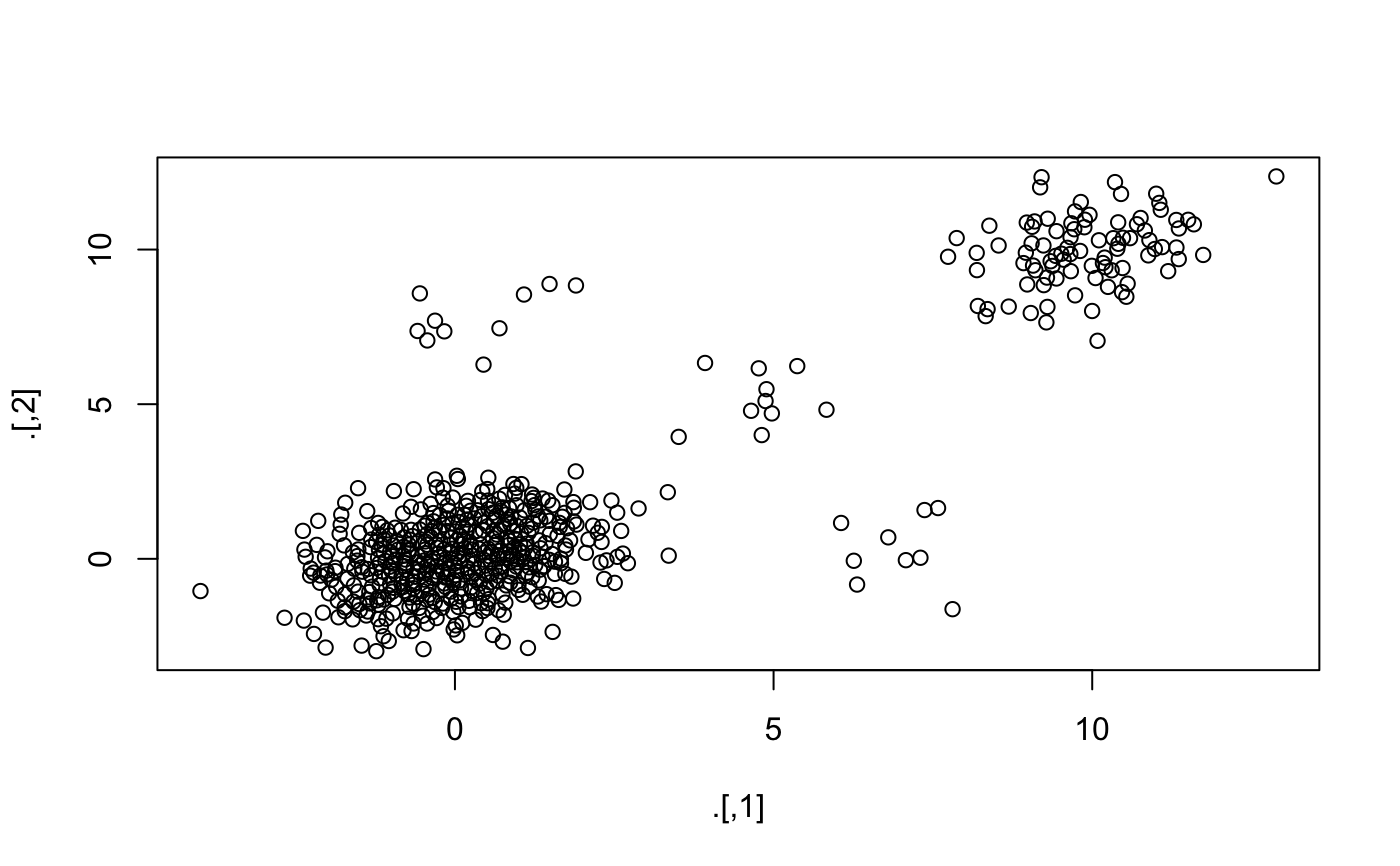
library("magrittr") dataset <- rbind( MASS::mvrnorm(1000, mu = c(0, 0), Sigma = matrix(c(1, 0.3, 0.3, 1), 2)) , MASS::mvrnorm(100, mu = c(10, 10), Sigma = matrix(c(1, 0.3, 0.3, 1), 2)) , MASS::mvrnorm(10, mu = c(5, 5), Sigma = matrix(c(1, 0.3, 0.3, 1), 2)) , MASS::mvrnorm(10, mu = c(0, 8), Sigma = matrix(c(1, 0.3, 0.3, 1), 2)) , MASS::mvrnorm(10, mu = c(7, 0), Sigma = matrix(c(1, 0.3, 0.3, 1), 2)) ) plot(dataset)#> Min. 1st Qu. Median Mean 3rd Qu. Max. #> 0.02362 0.07265 0.10334 0.16972 0.16889 1.99385